Sitemap
A list of all the posts and pages found on the site. For you robots out there, there is an XML version available for digesting as well.
Pages
Posts
Future Blog Post
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Blog Post number 4
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 3
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 2
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 1
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
portfolio
Distributed Systems Lab
This repository contains my Go implementations of distributed systems primitives from MIT 6.5840: Distributed Systems. Projects include a simplified MapReduce framework with task scheduling, worker registration, fault tolerance, and coordination between mappers and reducers for parallel data processing; a full Raft consensus algorithm implementation covering leader election, log replication, term persistence, and crash recovery; a replicated key/value store layered on Raft that guarantees linearizability under failures; a sharded key/value service that supports dynamic configuration and shard migration between replica groups; and a persistent key/value store with snapshotting and state recovery to handle large logs and node restarts. Each module is built with a focus on fault tolerance, concurrency, and distributed correctness under real-world conditions. Github repository 
Golong Distributed Cache
A lightweight, in-memory distributed caching system implemented in Go, designed for scalability and simplicity. This project is a distributed in-memory key-value cache built in Go, designed to demonstrate core principles of scalable system design. It uses gRPC for communication between cache nodes and supports basic operations like Get, Set, and Delete. Keys are distributed across nodes using consistent hashing, which ensures load balancing and minimizes data movement when nodes are added or removed. Each node includes an LRU-based local cache and acts as both a server and client, forwarding requests to the correct node if needed. An HTTP gateway provides a simple REST API interface for users, internally routing requests via gRPC. The system is modular and can be extended with features like persistence or replication. This project is ideal for understanding distributed coordination, request routing, and cache eviction strategies using Go and gRPC. It includes unit tests and documentation to help developers quickly run and experiment with a multi-node setup. Github repository
Golong Distributed File System
A lightweight, peer-to-peer distributed file system implemented in Go. This project demonstrates the core principles of distributed storage, including file replication, peer discovery, and secure data transfer. Peer-to-Peer Architecture: Nodes communicate directly, eliminating the need for a central server. File Replication: Ensures data redundancy by replicating files across multiple peers. Secure Communication: Utilizes cryptographic techniques to secure data transmission between nodes. Modular Design: Components like storage, networking, and cryptography are modular, facilitating easy maintenance and extension. Command-Line Interface: Simple CLI for interacting with the file system. Github repository 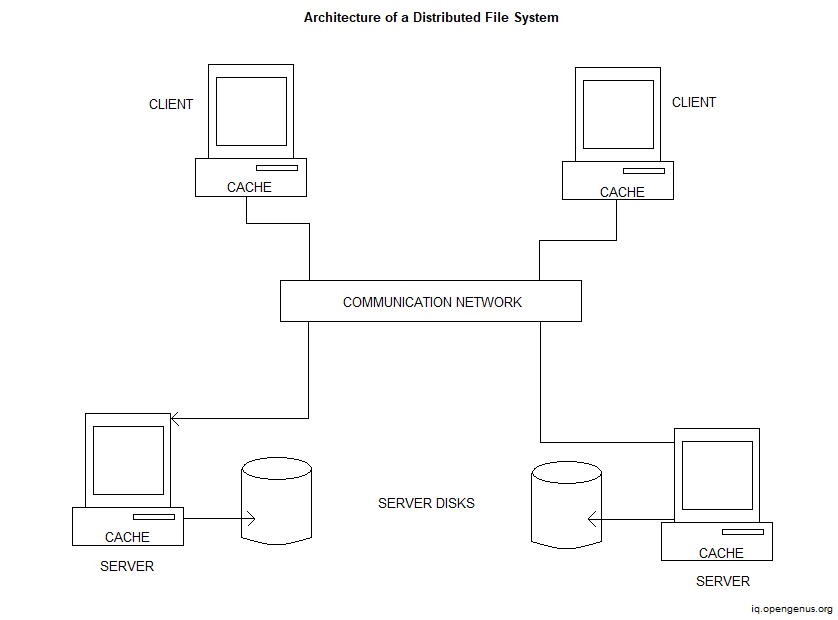
Backtrack
A biophysics tool for tracking cell motion and division. Backtrack uses Deep learning-based segmentation and motion tracking (MOT) algorithms. Integrated SSN (Spatiotemporal Sequence Networks) for improved cell tracking and segmentation of E. coli microscope data, optimizing tracking accuracy and performance. Optimized deep learning model performance for biological datasets using PyTorch, NumPy, OpenCV, scikit-image. Bactrack is inspired by ultrack. Bactrack uses segmentation hierarchy to allow various segmentation scenarios, and hierarchy is built on Omnipose dynamic/pixel clustering logic, and using MIP solver to assign cell from frame to frame by maximize weights. For assignment algorithm, Bactrack includes following MIP solvers: HiGHS, CBC, Gurobi for tracking assignment task. All of these MIP solver will return the same optimized global maximum result but with different run-time speed. For performance comparsion between MIP solvers check this benchmark. In short, the speed of Gurobi is the fastest (Gurobi > HiGHS > CBC). Github repository 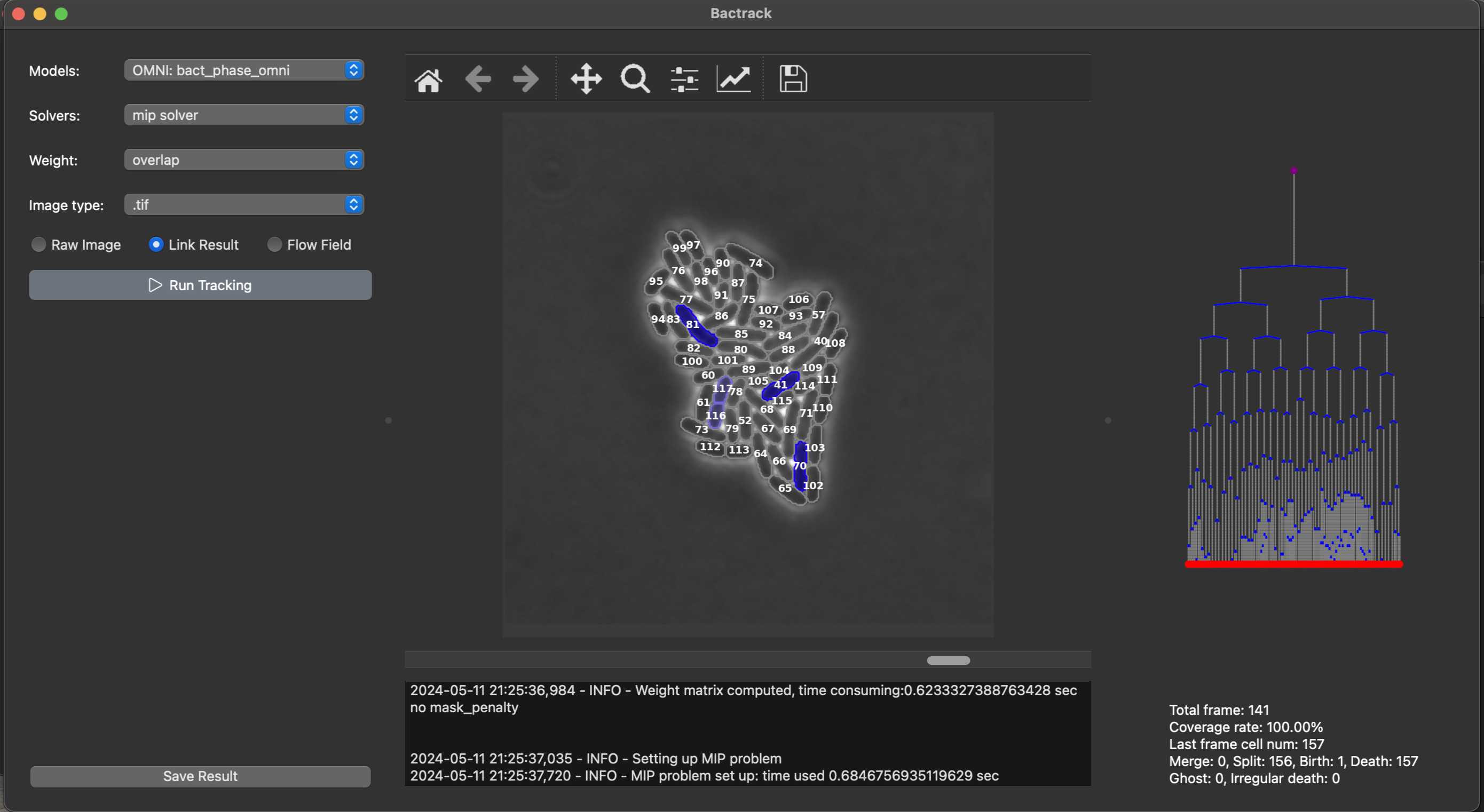
Precovery
Precovery of planetoids is a census of minor planets in by optimize condense data searching algorithm. Utilized Python, SQL, bash, and Git to process raw data and enhance the ADAM database. Expanded the database by 20%, adding over 500 million objects and 5 billion observations. Precovery returns observations that lie within the angular tolerance of the predicted location of an input orbit propagated and mapped to the indexed observations. These observations are termed PrecoveryCandidates. Optionally, precovery can also return FrameCandidates which are frames where the orbit intersected the Healpix-mapped exposure for a specific dataset but no observations were found within the angular tolerance. In this case, quantities specific to individual observations will be returned as NaNs (mjd, ra_deg, dec_sigma_arcsec, ra_sigma_arcsec, mag, mag_sigma, observation_id, delta_ra_arcsec, delta_dec_arcsec, distance_arcsec), with the remaining quantities that define the Healpix-mapped exposure returned as normal Github repository 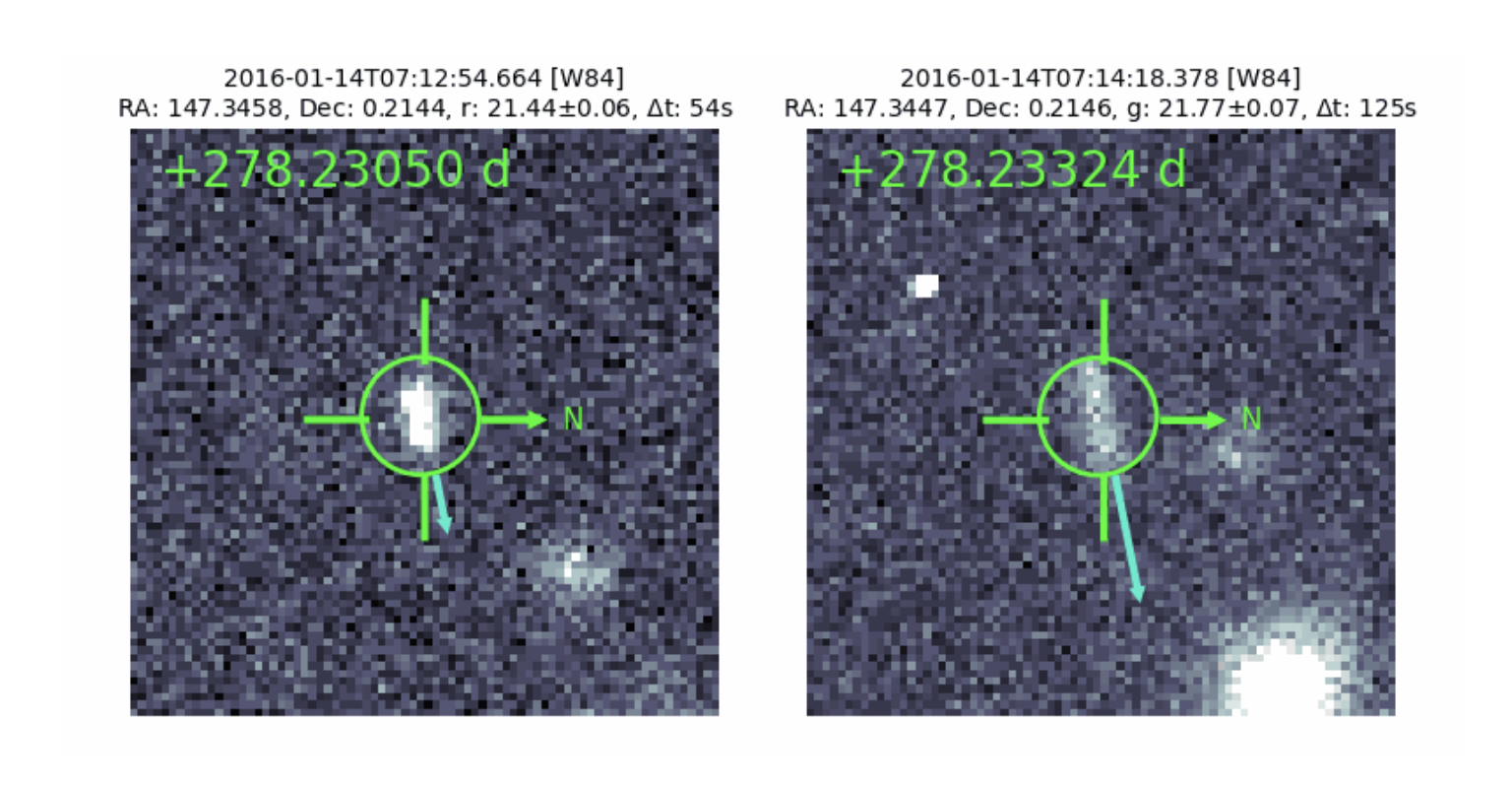
publications
Paper Title Number 1
Published in Journal 1, 2009
This paper is about the number 1. The number 2 is left for future work.
Recommended citation: Your Name, You. (2009). "Paper Title Number 1." Journal 1. 1(1).
Download Paper | Download Slides | Download Bibtex
Paper Title Number 2
Published in Journal 1, 2010
This paper is about the number 2. The number 3 is left for future work.
Recommended citation: Your Name, You. (2010). "Paper Title Number 2." Journal 1. 1(2).
Download Paper | Download Slides
Paper Title Number 3
Published in Journal 1, 2015
This paper is about the number 3. The number 4 is left for future work.
Recommended citation: Your Name, You. (2015). "Paper Title Number 3." Journal 1. 1(3).
Download Paper | Download Slides
Paper Title Number 4
Published in GitHub Journal of Bugs, 2024
This paper is about fixing template issue #693.
Recommended citation: Your Name, You. (2024). "Paper Title Number 3." GitHub Journal of Bugs. 1(3).
Download Paper
talks
Talk 1 on Relevant Topic in Your Field
Published:
This is a description of your talk, which is a markdown file that can be all markdown-ified like any other post. Yay markdown!
Conference Proceeding talk 3 on Relevant Topic in Your Field
Published:
This is a description of your conference proceedings talk, note the different field in type. You can put anything in this field.
teaching
Teaching experience 1
Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Teaching experience 2
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.

